New Irish study reveals how vancomycin-resistant Enterococcus faecium, a globally emerging hospital superbug, acquires and rapidly transfers resistance to antibiotics of last resort
Infections caused by antibiotic resistant bacteria are a leading threat to public health worldwide. The bacterium Enterococcus faecium has emerged globally as a significant cause of hospital-acquired infections because of its naturally low susceptibility to many commonly used antibiotics. Therapeutic options are limited to a small number of antibiotics including vancomycin. However, rising proportions of vancomycin-resistant E. faecium (VREfm) infections have been reported worldwide with limited treatment options. Data from the European Centre for Disease Prevention and Control revealed that for over a decade, the Republic of Ireland has reported one of the highest rates of invasive VREfm infections in Europe, ranging between 32.5%-45.8% (2006-2019).
Microbiologists at the Dublin Dental University Hospital (DDUH), Trinity College, the Irish National MRSA Reference Laboratory at St. James’s Hospital (SJH) Dublin, the Department of Clinical Microbiology, SJH and the Department of Clinical Microbiology, Hvidovre University Hospital, Denmark used whole-genome sequencing to investigate 600 VREfm from Irish hospitals in comparison with 1835 international isolates from 30 countries.
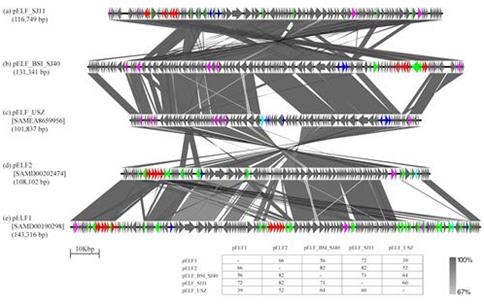
Irish VREfm were found to be genetically diverse yet distinctly different to VREfm from other countries. Irish VREfm have evolved independently and harbour a unique genetic transposon element that can mobilize vancomycin resistance genes into vancomycin-susceptible isolates of diverse genetic lineages using a combination of circular and linear plasmids and insertion sequences, thus giving rise to new strains of VREfm.
Commenting on the significance of the findings, Professor David Coleman from the DDUH Division of Oral Biosciences (https://www.tcd.ie/dental/people/dcoleman/) said: “this research is a compelling example of how new genetic technologies such as high-throughput whole-genome sequencing can be used to reveal new and important information about antibiotic-resistant bacteria, so we may better understand, and ultimately prevent and treat life-threatening infections.
This study was funded by Health Research Board Grant ILP-POR-2019-010 and published in the international journal Journal of Antimicrobial Chemotherapy
https://academic.oup.com/jac/advance-article/doi/10.1093/jac/dkab393/6423109